Abstract
Recently, there has been a surge of interest in extending the success of large language models (LLMs) from texts to molecules. Most existing approaches adopt a graph neural network to represent a molecule as a series of node tokens for molecule-language alignment, which, however, have overlooked the inherent hierarchical structures in molecules. Notably, highe-rorder molecular structures contain rich semantics of functional groups, which encode crucial biochemical functionalities of the molecules. We show that neglecting the hierarchical information in tokenization will lead to subpar molecule-language alignment and severe hallucination. To address this limitation, we propose HIerarchical GrapH Tokenization (HIGHT). HIGHT employs a hierarchical graph tokenizer that encodes the hierarchy of atom, motif, and molecular levels of informative tokens to improve the molecular perception of LLMs. HIGHT also adopts an augmented instruction tuning dataset, enriched with the hierarchical graph information, to further enhance the molecule-language alignment. Extensive experiments on 14 real-world benchmarks verify the effectiveness of HIGHT in reducing hallucination by 40%, and significant improvements in various molecule-language downstream tasks. The project is available at https: //higraphllm.github.io/.
HIGHT Framework
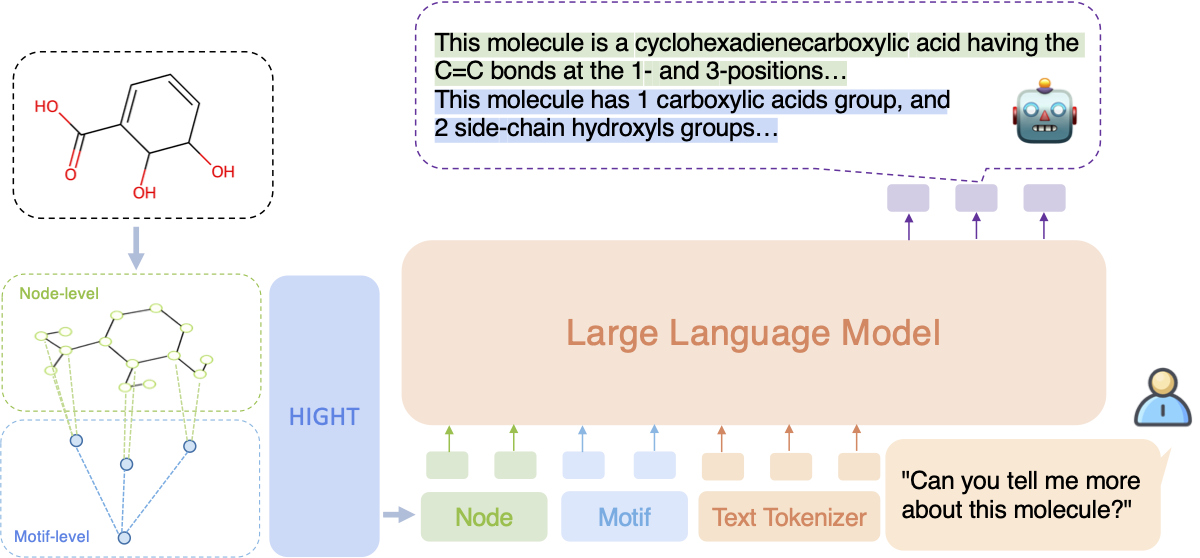
Figure 1. Illustration of the HIGHT framework.
Motif Hallucination
With only node-level tokens (i.e., discrete atom embeddings), LLMs have to identify and connect the nodes within a specific functional group in order to align useful molecular structures in a molecule to the corresponding language descriptions. Yet, due to the arbitrary order of atoms and position biases in LLMs, it is harder to distinguish each functional group, which further leads to hallucination and subpar alignment.
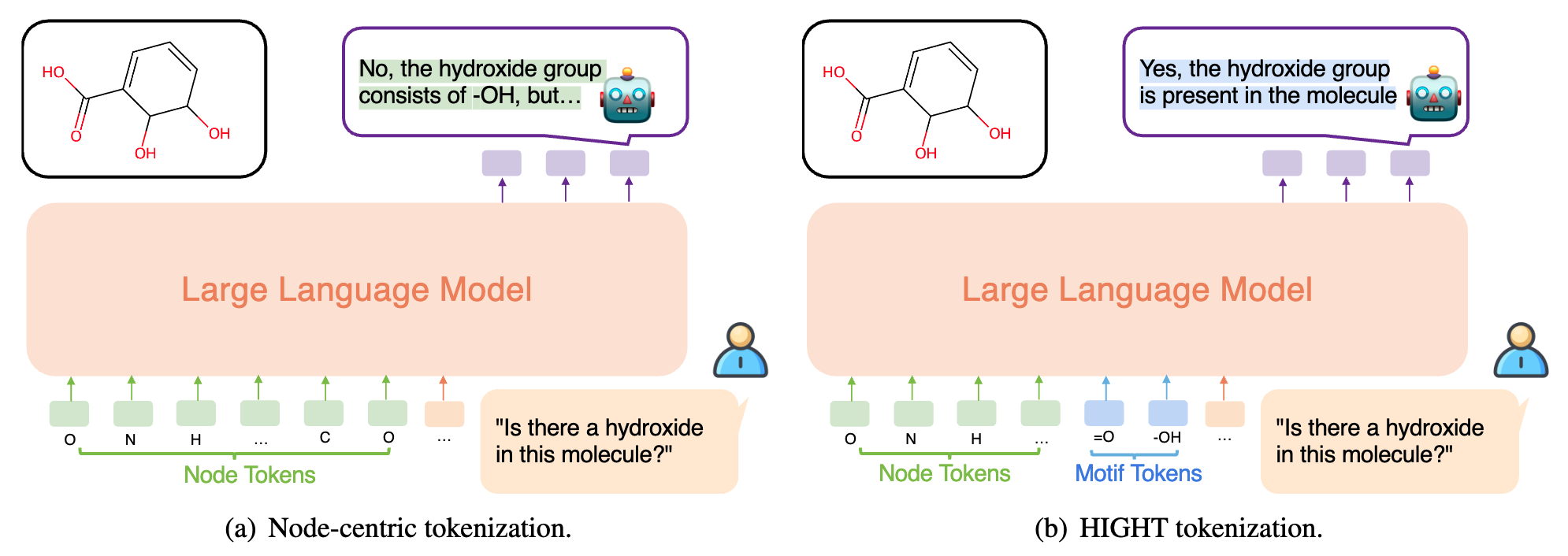
Figure 2. Illustration of Motif Hallucinations.
To understand the issue of node-centric tokenization more clearly, we construct a simple benchmark called MotifHallu, which measures the hallucination of common functional groups by LGLMs. Specifically, we consider the 38 common functional groups in RDKit, and construct 23,924 examples of query-answer pairs that asks LLMs about the existence of the functional groups.
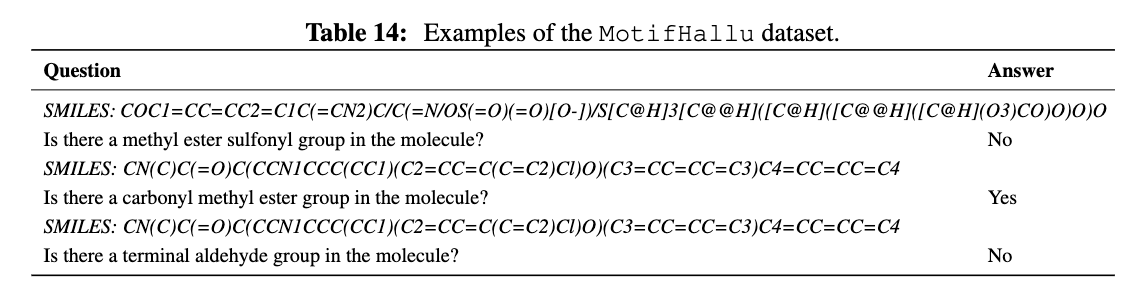
Figure 3. Examples of MotifHallu benchmark.
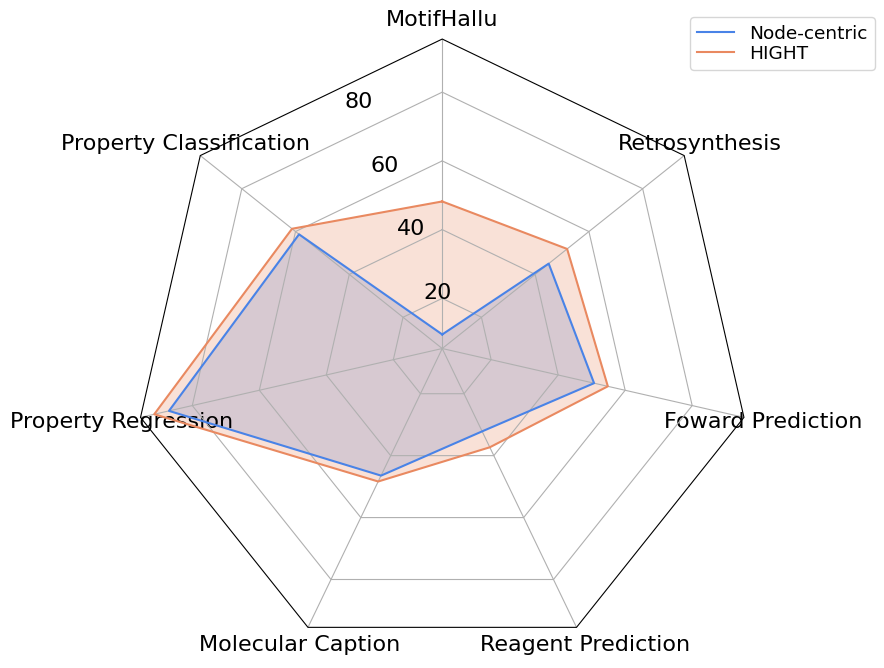
Due to the limitation of motif perception and suboptimal alignment, it can be observed a severe hallucination ratio. In contrast, HIGHT significantly alleviates the hallucination.
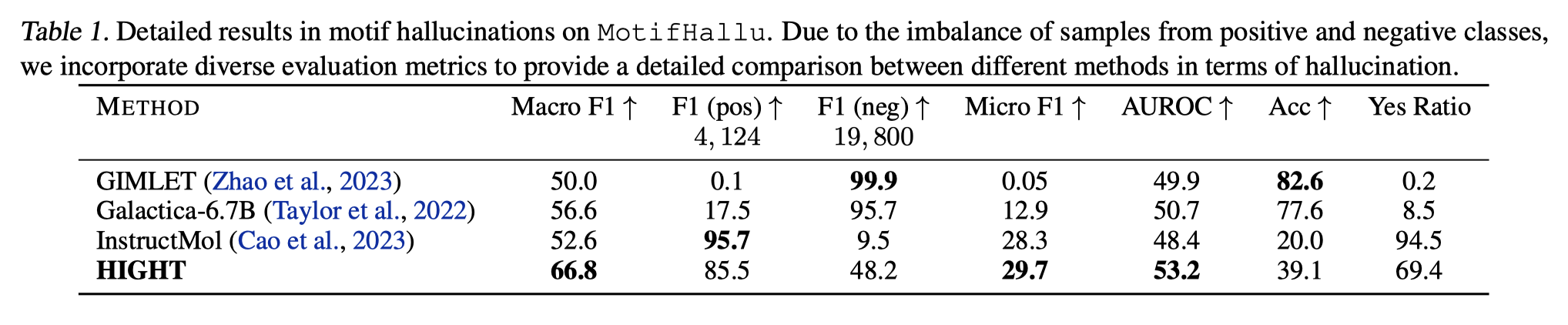
Figure 4. Detailed results in motif hallucinations on MotifHallu. Due to the imbalance of samples from positive and negative classes, we incorporate diverse evaluation metrics to provide a detailed comparison between different methods in terms of hallucination.
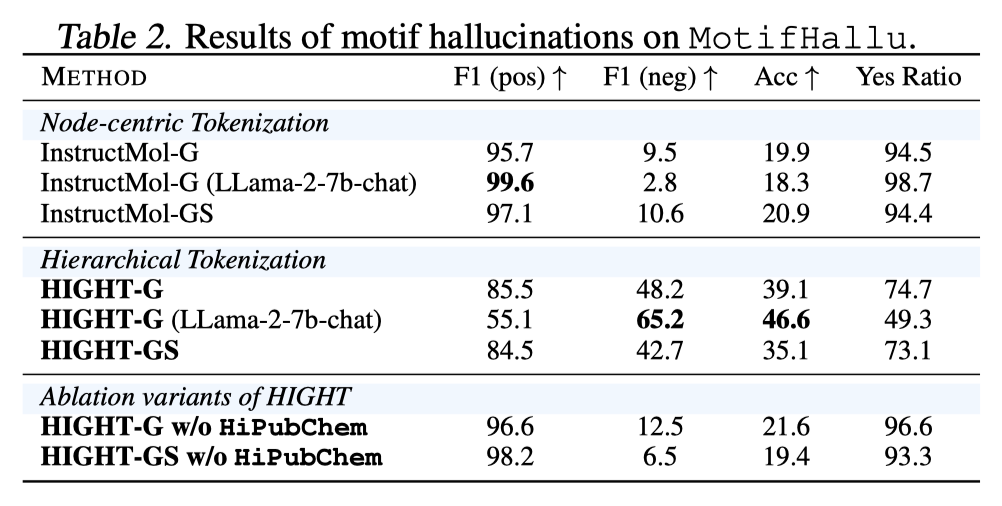
Figure 5. Influence of tokenization on motif hallucinations on MotifHallu.
Results on Downstream Tasks
We also compare the performances of HIGHT with other LGLMs across 6 downstream tasks including including property prediction, molecular description, and chemical reaction prediction.
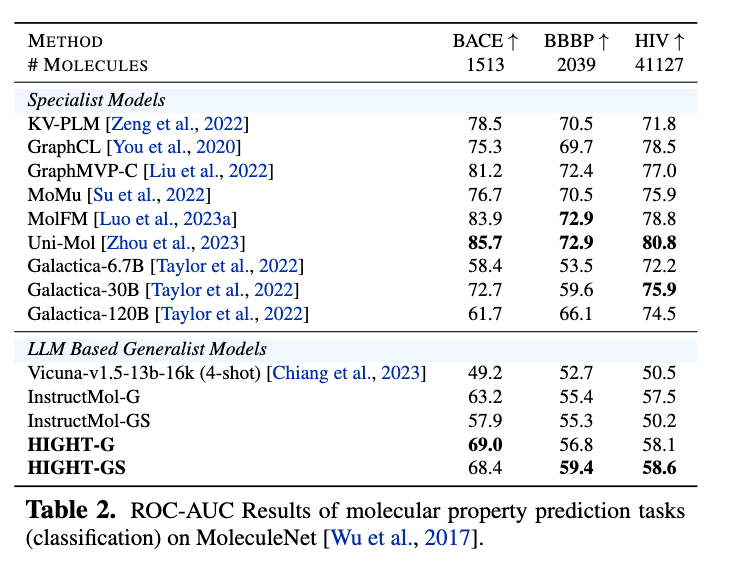
Figure 6. ROC-AUC Results of molecular property prediction tasks (classification) on MoleculeNet. Evaluation on InstructMol and HIGHT adopt the likelihood of the tokens of ``Yes'' and ``No''. Most of the instruction tuning datasets are from GIMLET. SIDER and ClinTox are converted following the MoleculeNet task description.
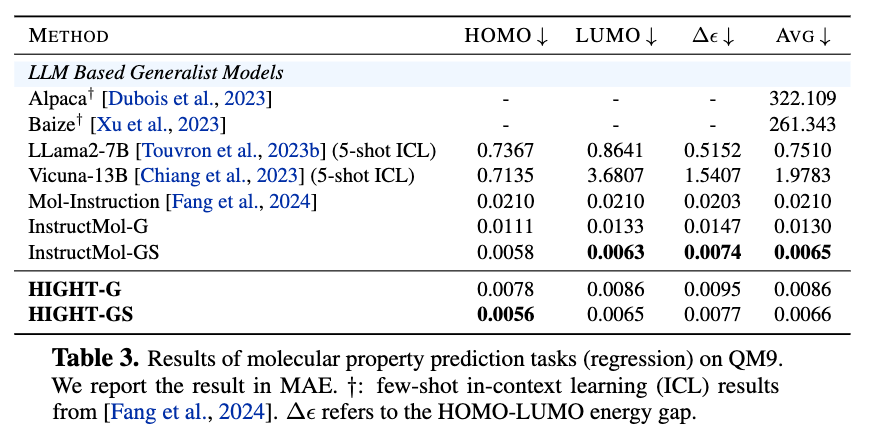
Figure 7. Results of molecular property prediction tasks (regression) on QM9.
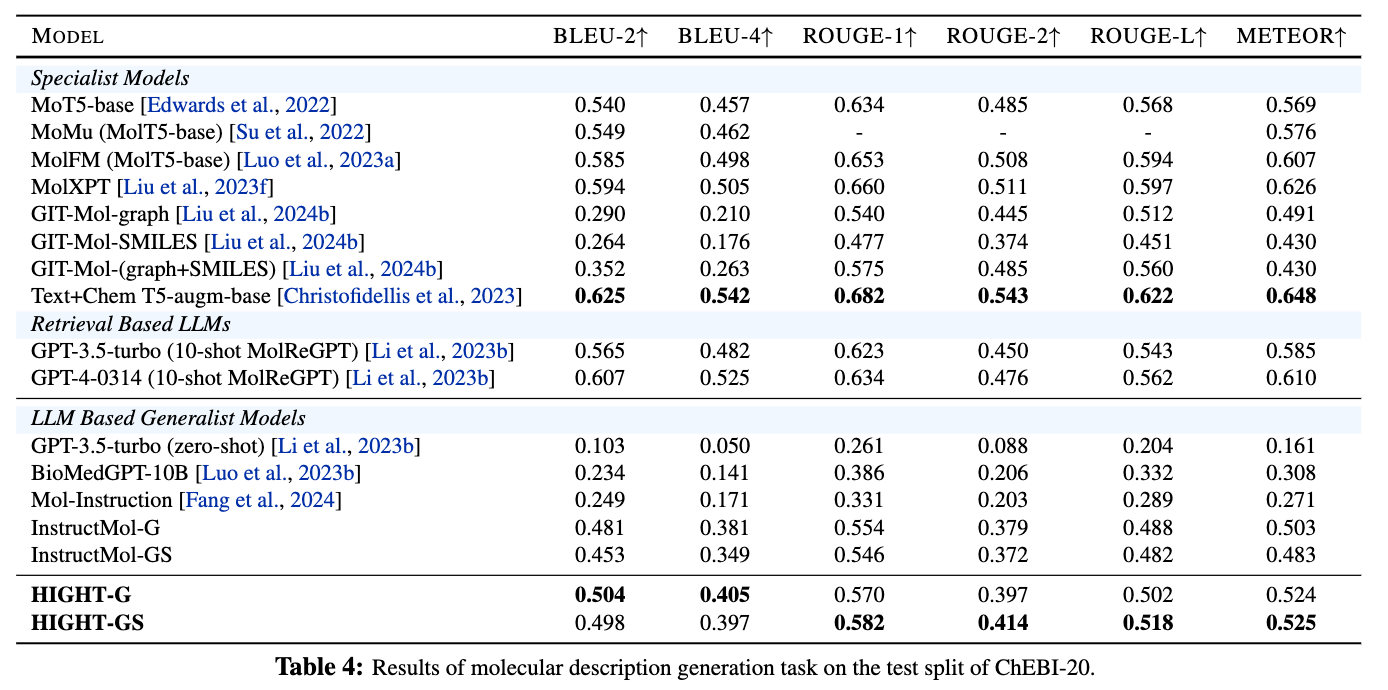
Figure 8. Results of molecular description generation task on the test split of ChEBI-20.
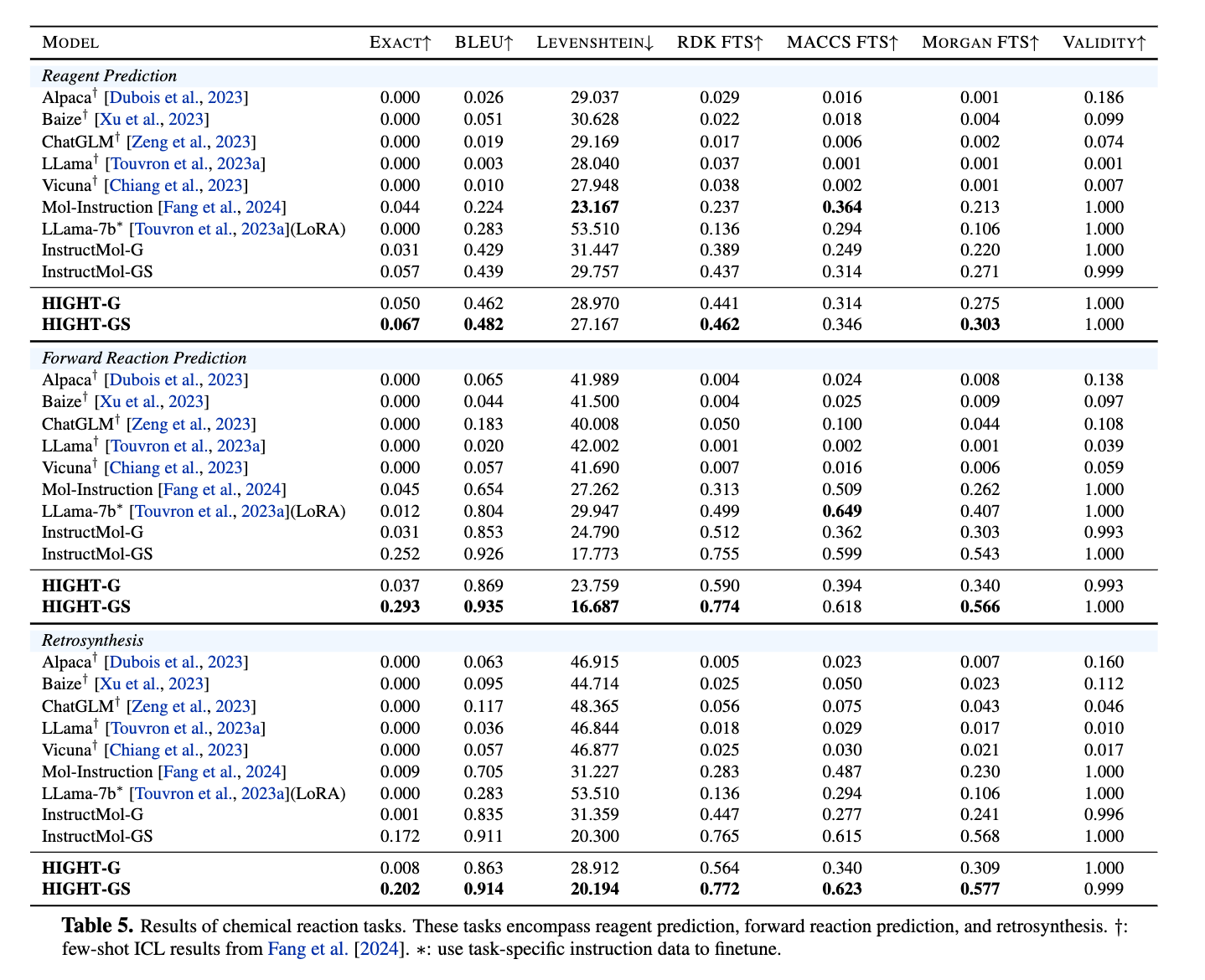
Figure 9. Results of chemical reaction tasks from Mol-Instructions.
We follow the previous practice in training and evaluating generalist models and consider the two settings: a) As shown in Fig. 3(a), we first train the model with all chemical reaction prediction data by 3 epochs to elicit the format following and the knowledge adaption capabilities of the LGLMs after stage 1. The models are named with “-All”; b) As shown in Fig. 3(b), we train the model with retrosynthesis task data and evaluate the zero-shot transfer performance on forward reaction prediction. Under both settings, we can find that HIGHT boosts the generalist capabilities significantly. To better understand the effectiveness of distinct components in HIGHT, we conduct ablation studies that train InstructMol with the laplacian positional encodings or with HiPubChem, as given in Fig. 3(c). We can find that, merely incorporating positional encoding or hierarchical instruction tuning is not sufficient to achieve the same performance as HIGHT. On the contrary, without a proper architecture design as HIGHT, LGLMs with previous node-centric tokenization with HiPubChem will confuse LLMs and even lead to degenerated downstream task performances. In addition, we also compare LGLMs with llama2 backbone. As shown in Fig. 3(a), HIGHT still significantly boosts the performance.
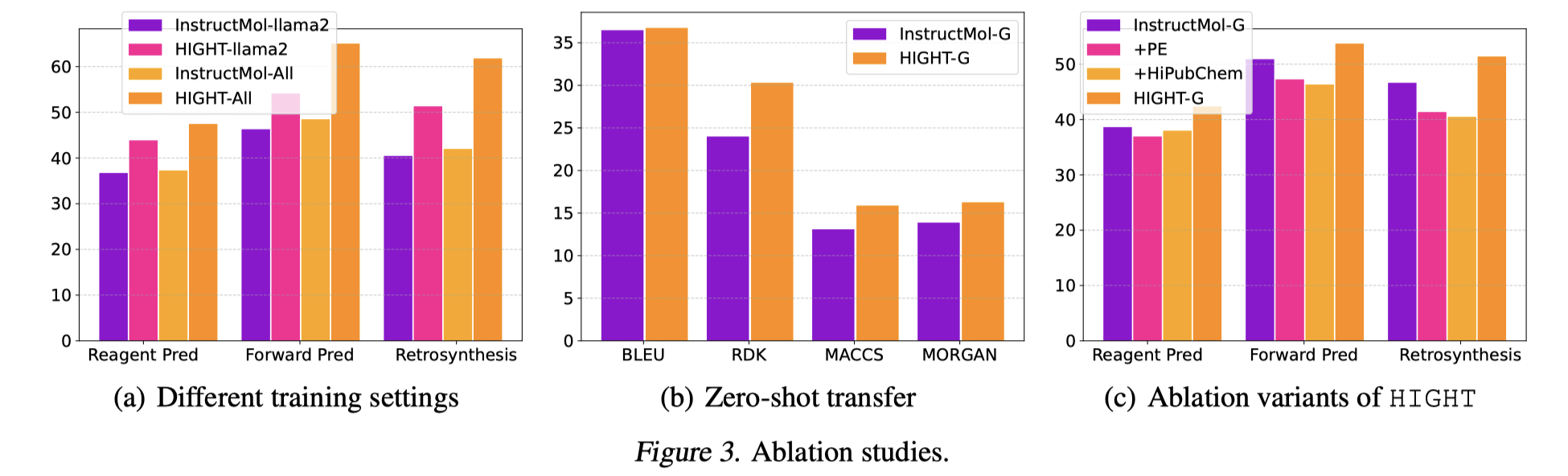
Figure 10. Ablation studies.
Contact
Welcome to check our paper for more details of the research work. If there is any question, please feel free to contact us.
If you find our paper and repo useful, please consider to cite:
@article{higraphllm2024,
title={HIGHT: Hierarchical Graph Tokenization for Graph-Language Alignment},
author={Yongqiang Chen and Quanming Yao and Juzheng Zhang and James Cheng and Yatao Bian},
year={2024},
journal = {arXiv preprint},
volume = {arXiv:2406.14021}
}